Swiss Data Aggregator DAGI FAQs
Data preparation
Do I have to upload my entire database into the Data Aggregator?
There is no need to upload all of your database into the Data Aggregator DAGI. You can choose to upload only the most important fields for a selected set of records. Keep in mind that the key element of the data you import in DAGI is the catalogNumber attribute, which has to be unique for all of your records. If a given catalogNumber value does not yet exist in your DAGI Collection, then it is created when importing a dataset. If a given catalogNumber value already exists in the DAGI Dataset, then its attributes (other fields) are simply updated when importing a new file.
To help you select your fields, here is a table with the most important Darwin Core terms and an example line. You can use it to organise your dataset for the upload into DAGI.
| scientificName | acceptedNameUsage | family | basisOfRecord | partOfOrganism | catalogNumber | recordedBy | recordedByID | recordNumber | verbatimEventDate | day | month | year | end_of_period_day | end_of_period_month | end_of_period_year | eventDate | continent | higherGeography | country | countryCode | stateProvince | county | locality | decimalLatitude | decimalLongitude | geodeticDatum | coordinateUncertaintyInMeters | verbatimElevation | identifiedBy | identifiedByID | rightsHolder | preparations | typeStatus | yearCollectionEntrance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pinus picea L. | Abies alba Mill. | Pinaceae | preservedSpecimen | plant tissue | inventory-1234 | Weber Morgan | 0000-0002-1043-7587 | MW-54 | 6/2024 | 01 | 06 | 2024 | 30 | 06 | 2024 | 2024-06-01/2024-06-30 | Europa | Alpen | Switzerland | CH | Bern | Interlaken-Oberhasli (administrative district) | Luuswald | 46.701815 | 7.971722 | WGS84 | 500 | 1050-1120 m | Weber Morgan | 0009-0000-0012-XXXX | Herbarium X | dried plant | 2015 |
The Darwin Core Github repository also offers files with all or a selection of the Darwin Core terms : Github tdwg/dwc/dist
How does the update of my data in the Data Aggregator work?
You can update your data in DAGI by importing a new import file. This file must have the two mandatory fields (catalogNumber and scientificName). The other fields in the file can be either the same as previously imported or only the fields that have to be updated. It is up to you.
During the new import, DAGI checks the catalogNumber value to determine if a record is already present in the DAGI dataset, or if it is newly imported.
- When the record is already present, all other attributes imported are updated (scientificName too).
- When the record is new (new catalogNumber), the record is added to the records table with all imported attributes.
How does the update of my data on GBIF work?
You can update your data on GBIF by publishing your dataset again.
After the first publication, DAGI retrieves the datasetID that has been automatically generated by GBIF for the new dataset. During the new publication, DAGI checks if the datasetID exists on GBIF. It then updates all of the records and metadata of the GBIF dataset with the Darwin Core Archive file it prepared.
But my database/dataset is not formatted in Darwin Core, do I have to change everything?
Rest assured, you do not need to change your database/dataset dramatically. The most important thing is to find the easiest and fastest way to adapt your database/dataset to import it in DAGI. Here are our 3 most popular suggestions:
| Barcode | catalogNumber | Species | scientificName | ... |
|---|---|---|---|---|
| XXX-0123456 | XXX-0123456 | Cyclamen hederifolium | Cyclamen hederifolium Aiton | ... |
| XXX-7891011 | XXX-7891011 | C. hederifolium | Cyclamen hederifolium Aiton | ... |
✅ No changes of original columns/fields
❌ Duplicated in multiple columns
❌ If not dynamic, then mistakes can lower the dataset/database quality
catalogNumber |
scientificName |
... | eventDate |
|---|---|---|---|
| XXX-0123456 | Cyclamen hederifolium Aiton | ... | 1905-08-12 |
| XXX-7891011 | Cyclamen hederifolium Aiton |
... | 1968-06-12 |
✅ No more changes in the future
❌ Difficult to change habits regarding field names
❌ Needs a deep cleaning of the whole database/dataset
Your original data
| Barcode | Species | Date of collect | Storage room | ... |
|---|---|---|---|---|
| XXX-0123456 | Cyclamen hederifolium | 12 VIII 1905 | General collection | ... |
| XXX-7891011 | C. hederifolium | 23.6.68 | Regional collection | ... |
+
Table imported in the Aggregator
| catalogNumber | scientificName | eventDate |
|---|---|---|
| XXX-0123456 | Cyclamen hederifolium Aiton | 1905-08-12 |
| XXX-7891011 | Cyclamen hederifolium Aiton | 1968-06-23 |
✅ Full control of the data you share
❌ Duplicated data
❌ Extensive preparation work for every update of the data online
Which fields are required/mandatory?
Minimal mandatory fields of the Data Aggregator
| DwC term | DwC definition | In most databases | Examples |
|---|---|---|---|
| scientificName | The full scientific name, with authorship and date information if known, or the name in lowest level taxonomic rank that can be determined. | Scientific name nom scientifique Wissenschaftliche Name Full name Nom complet |
Cyclamen hederifolium Aiton Vulpes vulpes (Linnaeus, 1758) |
| catalogNumber | A unique identifier for the record within the data set or collection. | Code-barre Numéro Barcode Nummer Numéro d’inventaire |
G00009201 Sheet-2765149 |
Fields in the Data Aggregator with special values required
The DAGI’s available attributes are based mainly on the Darwin Core terms but there are also supplementary attributes from the GBIF extensions, ABCD standard and GBIF-CH. The use of some of these attributes is also specific to our national installations.
| DAGI Attribute | Value required | Examples |
|---|---|---|
| catalogNumber | Your institution unique ID for a specimen (ideally starting with your institution or collection code) | G00547679 |
| occurrenceID | Info Species data center unique ID for a specimen already published on GBIF before 2025 | NISM-BRYO-537533 |
| materialSampleID | GBIFCH unique ID for a specimen | GBIFCH000014 |
| gbifCHID | GBIFCH unique ID for a specimen | GBIFCH000014 |
| swissCoordinatesLv95_E | Swiss longitude coordinate in CH1903+/LV95 format _(always starts with a "2")_ |
2598633.94 |
| swissCoordinatesLv95_N | Swiss latitude coordinate in CH1903+/LV95 format _(always starts with a "1")_ |
1200386.85 |
| swissCoordinatesLv03_E | Swiss longitude coordinate in CH1903/LV03 format _(always bigger than the latitude coordinate)_ |
657499.41 |
| swissCoordinatesLv03_N | Swiss latitude coordinate in CH1903/LV03 format _(always smaller than the longitude coordinate)_ |
191750.14 |
| associatedMedia | empty OR the URL to a public deposit of your specimen image | https://www.digitalis.uzh.ch/media/specimen/293/Z-000293332.jpg |
Additional fields increasing data quality in the Data Aggregator (MIDS)
The MIDS is the Minimum Information about a Digital Specimen. The four levels of MIDS (0, 1, 2, 3) have been implemented in DAGI in order to provide a visualisation of the degree of information associated to a record. The levels are interconnected: all attributes of a certain level have to be informed in order to reach the next level.
MIDS 0 : Bare - A bare or skeletal record making the association between an identifier of a physical specimen and its digital representation, allowing for unambiguous attachment of all other information.
MIDS 1 : Basic - A basic record of specimen information enabling basic discoverability as well as how the user is permitted to use the data.
MIDS 2 : Intermediate - A regular level of information including data that have been agreed over time as essential for most scientific purposes.
MIDS 3 : Extended - An extended level of information about a specimen including identifiers enabling connections to be made to other data present or known about the specimen.
| MIDS | DAGI Attribute | Definition | Comment |
|---|---|---|---|
| 0 | partOfOrganism | Part or parts of the organism that have been preserved, e.g. shell, skeleton, skull, soft tissue. |
This is not a Darwin Core term, but it is available in DAGI. It is possible to concatenate multiple values with the use of the vertical bar " | ". |
| taxonID | An identifier for the set of dwc:Taxon information. May be a global unique identifier or an identifier specific to the data set. | By encoding your records, you automatically obtain the taxonID of GBIF | |
| 1 | eventDate | The date-time or interval during which a dwc:Event occurred. For occurrences, this is the date-time when the dwc:Event was recorded. Not suitable for a time in a geological context. | The structure must follow the ISO 8601-1:2019 standard. 2025-04-08 2025-04-08/2025-04-10 |
| recordedBy | A list (concatenated and separated) of names of people, groups, or organizations responsible for recording the original Occurrence. The primary collector or observer, especially one who applies a personal identifier (recordNumber), should be listed first. | ||
| typeStatus | A list (concatenated and separated) of nomenclatural types (type status, typified scientific name, publication) applied to the subject. | Good practice on GBIF is to leave it empty if it is not a Typus. | |
| originalNameUsage | The taxon name, with authorship and date information if known, as it originally appeared when first established under the rules of the associated dwc:nomenclaturalCode. The basionym (botany) or basonym (bacteriology) of the dwc:scientificName or the senior/earlier homonym for replaced names. | ||
| continent | The name of the continent in which the Location occurs. | ||
| country | The name of the country or major administrative unit in which the Location occurs. | Best practice is to use current existing country names and not historical countries. | |
| stateProvince | The name of the next smaller administrative region than country (state, province, canton, department, region, etc.) in which the Location occurs. | ||
| county | The full, unabbreviated name of the next smaller administrative region than stateProvince (county, shire, department, etc.) in which the Location occurs. | ||
| higherGeography | A list (concatenated and separated) of geographic names less specific than the information captured in the locality term. | ||
| locality | The specific description of the place. | ||
| decimalLatitude | The geographic latitude (in decimal degrees, using the spatial reference system given in geodeticDatum) of the geographic center of a Location. Positive values are north of the Equator, negative values are south of it. Legal values lie between -90 and 90, inclusive. | A coordinate conversion in DAGI allows to import the swiss coordinates and obtain the decimalLatitude by encoding. | |
| decimalLongitude | The geographic longitude (in decimal degrees, using the spatial reference system given in geodeticDatum) of the geographic center of a Location. Positive values are east of the Greenwich Meridian, negative values are west of it. Legal values lie between -180 and 180, inclusive. | A coordinate conversion in DAGI allows to import the swiss coordinates and obtain the decimalLongitude by encoding. | |
| verbatimDepth | The original description of the depth below the local surface. | ||
| verbatimElevation | The original description of the elevation (altitude, usually above sea level) of the Location. | ||
| yearCollectionEntrance | The four-digit year of collection entrance of a specimen (earliest year of occurrence in absence of a documented collection event). | This is not a Darwin Core term, but it is available in DAGI. | |
| occurrenceID | An identifier for the dwc:Occurrence (as opposed to a particular digital record of the dwc:Occurrence). In the absence of a persistent global unique identifier, construct one from a combination of identifiers in the record that will most closely make the dwc:occurrenceID globally unique. | If not informed during import, this attribute is automatically copied from the catalogNumber. | |
| 2 | verbatimEventDate | The verbatim original representation of the date and time information for a dwc:Event. | |
| identifiedBy | A list (concatenated and separated) of names of people, groups, or organizations who assigned the dwc:Taxon to the subject. | ||
| identificationQualifier | A brief phrase or a standard term ("cf.", "aff.") to express the determiner's doubts about the dwc:Identification. | ||
| identificationVerificationStatus | A categorical indicator of the extent to which the taxonomic identification has been verified to be correct. E.g. 0 (= "unverified" in HISPID/ABCD). |
||
| lastVerifiedBy | Person confirming the identification (usually a specialist of the corresponding systematic family). | This is not a Darwin Core term, but it is available in DAGI | |
| verbatimIdentification | A string representing the taxonomic identification as it appeared in the original record. | ||
| georeferencedBy | A list (concatenated and separated) of names of people, groups, or organizations who determined the georeference (spatial representation) for the dcterms:Location. | ||
| georeferenceVerificationStatus | A categorical description of the extent to which the georeference has been verified to represent the best possible spatial description for the dcterms:Location of the dwc:Occurrence. | ||
| verbatimCoordinates | The verbatim original spatial coordinates of the dcterms:Location. The coordinate ellipsoid, geodeticDatum, or full Spatial Reference System (SRS) for these coordinates should be stored in dwc:verbatimSRS and the coordinate system should be stored in dwc:verbatimCoordinateSystem. | ||
| verbatimLatitude | The verbatim original latitude of the dcterms:Location. The coordinate ellipsoid, geodeticDatum, or full Spatial Reference System (SRS) for these coordinates should be stored in dwc:verbatimSRS and the coordinate system should be stored in dwc:verbatimCoordinateSystem. | ||
| 3 | verbatimLongitude | The verbatim original longitude of the dcterms:Location. The coordinate ellipsoid, geodeticDatum, or full Spatial Reference System (SRS) for these coordinates should be stored in dwc:verbatimSRS and the coordinate system should be stored in dwc:verbatimCoordinateSystem. | |
| verbatimLocality | The original textual description of the place. | ||
| associatedMedia | URL to the original image deposited in a public access deposit | The URL can either be the URL to the institution's public image deposit or to the DAGI Media Store (Image Upload on DAGI). In order to be displayed on GBIF, the URL must be to the image itself and end with the extension .jpg/.jpeg/.png/etc. | |
| completeness | Degree of completeness of the specimen; may describe completeness of a part. e.g. complete, cephalon only, complete skull |
This is not a Darwin Core term, but it is available in DAGI | |
| otherCatalogNumbers | A list (concatenated and separated) of previous or alternate fully qualified catalog numbers or other human-used identifiers for the same dwc:Occurrence, whether in the current or any other data set or collection. | ||
| verbatimLabel | The content of this term should include no embellishments, prefixes, headers or other additions made to the text. Abbreviations must not be expanded and supposed misspellings must not be corrected. Lines or breakpoints between blocks of text that could be verified by seeing the original labels or images of them may be used. Examples of material entities include preserved specimens, fossil specimens, and material samples. Best practice is to use UTF-8 for all characters. Best practice is to add comment “verbatimLabel derived from human transcription” in dwc:occurrenceRemarks. |
Are there attributes in DAGI that are not part of Darwin Core?
Some of the attributes in DAGI have been borrowed from other sources (e.g. MIDS, GBIF Swiss Node) or have been homemade to answer specific needs of our swiss institutions (e.g. our swiss coordinate systems). These attributes can’t be yet published on GBIF (because GBIF does not have the structure for them). However some are used in DAGI during the encoding, and can ben sent to the Info Species data centers through the Validation.
Here is a table with these attributes and their definition:
| DAGI Attribute | Description | Examples |
|---|---|---|
| endOfPeriodDay | The integer day of a date marking the end of an interval in which the Event occurred. | 2, 30 |
| endOfPeriodMonth | The ordinal month of a date marking the end of an interval in which the Event occurred. | 2 (February), 11 (November) |
| endOfPeriodYear | The four-digit year of a date marking the end of an interval in which the Event occurred, according to the Common Era Calendar. | |
| generalEnvironment | Delarze et al. 2015: General Environment | |
| habitatCode | Habitat Code | 6.-2.1 |
| habitatContact | Delarze et al. 2015: Contact | |
| habitatInclusion | Delarze et al. 2015: Inclusion | |
| habitatRef | Delarze et al. 2015: Habitat Coding Reference | |
| influence | Delarze et al. 2015: Influence | |
| landscapeStructure | Delarze et al. 2015: Landscape Structure | |
| microStructure | Delarze et al. 2015: Microstructure | |
| substratum | Delarze et al. 2015: Substratum | |
| substratumState | Delarze et al. 2015: State of substratum | |
| placeOfOrigin | The place of origin for material that has been transported during its history e.g. glacial erratics and meteorites. | |
| evidenceType | Type of evidence or validation criterion considered (preferably according to a controlled vocabulary). | identified by genitalia |
| lastVerifiedBy | Person confirming the identification (usually a specialist of the corresponding systematic family). | Huber C. |
| lastVerifiedByID | Globally Unique Identifier of the person confirming the identification (usually a specialist of the coresponding systematic family). | https://orcid.org/0000-0003-3283-7764 |
| swissCoordinatesLv03_E | Swiss Coordinates CH1903/LV03, value towards the East (6 digits, https://epsg.io/21781). | 574175.61 |
| swissCoordinatesLv03_N | Swiss Coordinates CH1903/LV03, value towards the Nord (6 digits, https://epsg.io/21781). | 103975.67 |
| swissCoordinatesLv95_E | Swiss Coordinates CH1903+/LV95, value towards the East (7 digits, https://epsg.io/2056). | 574175.61 |
| swissCoordinatesLv95_N | Swiss Coordinates CH1903+/LV95, value towards the Nord (7 digits, https://epsg.io/2056). | 103975.67 |
| waterbodyID | The ID of the water body in which the Location occurs (according to a registry such as GEWISS). | CH0000180000 (for Walibach, Bennwil BL) |
| anatomicalDescription | Free text description of the preserved part of organism. | Mand. Dext. Mit Winkel und Ramus ascendens M3-P3 |
| articulation | Articulation in the preserved specimen - applies to invertebrate shells and exoskeletons as well as vertebrate skeletons. | articulated, dis-articulated, single valves |
| assemblageOrigin | The mode of origin of the assemblage. | unknown, allochthonous, autochthonous, paraautochthonous |
| barcodeLabel | Unique Specimen Identifier (Barcode Tag) | GBIFCH00376402, NMLU-ENT000115 |
| bioerosion | Damage due to biological action. | boring worms, sponges |
| completeness | Degree of completeness of the specimen; may describe completeness of a part. | complete, cephalon only, complete skull |
| depositionalEnvironmentText | Original environment in which the rock was deposited or the mineral formed. | hypersaline lagoon, lacustrine, intertidal |
| depositionalEnvironmentType | Keywords from enumerated list for indexing of depositional environments | |
| dnaBankID | Internal identifier assigned by the institution currently storing the DNA sample. | |
| dnaStableID | GBIFCH identifier assigned by the Biobank to the DNA sample. | |
| encrustation | Biological encrustations. | oysters and tube worms |
| extractionTemporaryID | Identifier assigned by the lab, temporarily ensuring links between genetic information. | |
| feedingPredationTraces | Aspects of feeding and predation. | ammonite with bite mark from plesiosaur, shell drilled by predatory gastropod |
| form | The original or a mold, cast etc. of the specimen. | |
| gbifDOI | GBIF DOI generated for a published dataset | |
| gbifCHID | GBIFCH unique identifier | |
| matrix | The sediment or mineral matrix enclosing the fossil. | |
| mineralization | The form of mineralization. | |
| organismQuantityMethod | Count type. Without indication, a number expressed in organismQuantity is interpreted as exact count. | exact count, estimation, minimum number |
| orientation | Orientation of the fossil remains in the host rock. | unknown, life position, topped |
| origColAuthor | Originator of a physical collection (“LEG”), possibly differing from the collector in the field (recordedBy). Information relevant for validation/plausibilisation of specimen occurrence records (cf. Monnerat et al. 2015). | |
| originalBiominerals | Origins of biomaterial preserved in the specimen. | |
| paleoCompleteness | An indication of the completeness of the representation of an organism. | disarticulated, complete |
| partOfOrganism | Part or parts of the organism that have been preserved. | shell, skeleton, skull, soft tissue |
| postBurialTransportation | Any post burial transport of fossil material. | river transport, scree slope |
| replacementMinerals | List of replacement minerals in the specimen. | silica |
| taphonomy | The life position, allochthonous death assemblage, post mortem history details etc. | |
| tissueBankID | GBIFCH identifier assigned by the Biobank to the tissue sample. | |
| yearCollectionEntrance | The four-digit year of collection entrance of a specimen (earliest year of occurrence in absence of a documented collection event). | 1897 |
| dnaBankInstitution | Biobank for long-term storage of DNA samples. | |
| dnaInstitution | Institution that still has the DNA. | |
| dnaStorageCode | Information on the place of storage of the DNA - Identifier, Location | |
| preservationAlterationText | Mineralogical changes in preserved specimens. | original shell material preserved, replacement minerals, re-crystallisation, silicification |
| preservationMethod | Preservation method for a specimen. | ethanol 70%, dried |
| preservationModeKeywords | Keywords for how fossil material has been preserved. | body, cast, mold, trace fossil, soft parts mineralised |
| preservationModeText | Mode of preservation. | is the specimen a cast or mold, are soft parts preserved or mineralised |
| preservationQuality | Preservation quality; includes preservation of anatomical detail and softparts. | poor, medium, good, excellent |
| preservationSpecialMode | Keywords for any special mode of preservation. | preserved in amber or frozen in tundra, tar pit |
| storageCode | The verbatim code of a storage or container unit; as it is mentioned or stated by the institution providing the resource. | |
| storageName | The type of storage that applies to entry. Usually refers to a vocabulary provided by the institution, e.g. “container”, “compactus”. | |
| tissueBankInstitution | Biobank for long-term storage of tissue samples. | |
| dateAvailable | Earliest release date conveined | |
| specificAuthorOfRecord | Author(s) to be cited for this record | |
| specifyEvent | Flag regulating the release of temporal data | |
| specifyLocality | Flag regulating the release of spatial data | |
| specifyOrganismName | Flag regulating the release of identification data | |
| specifyPerson | Flag regulating the release of person data |
Are there useful attributes that I never use in my database?
Sharing data can require information that have never been taken into consideration in a museum database simply because it is too obvious to specify. For instance, why have a partOfOrganism field in a collection database specialised in animal skulls or in fish fossils ? Well, from a FAIR perspective, this kind of information is important because it facilitates the filtering of data on one side and the analyses of a dataset on another side.
Here are some DAGI attributes that can be a very good addition to your import files or even your database:
| DAGI Attribute | Definition | Controlled vocabulary values examples |
|---|---|---|
| partOfOrganism | Part or parts of the organism that have been preserved | shell skeleton skull soft tissue whole plant leaf |
| degreeOfEstablishment | The degree to which a dwc:Organism survives, reproduces, and expands its range at the given place and time. | native captive cultivated released failing casual reproducing established colonising invasive widespreadInvasive |
| recordedByID | A list (concatenated and separated) of the globally unique identifier for the person, people, groups, or organizations responsible for recording the original dwc:Occurrence. (= recordedBy) | https://orcid.org/0000-0002-1825-0097 | https://orcid.org/0000-0002-1825-0098 |
| Adding a unique identifier to a name in a database is the best way to avoid confusions between homonymes. People registered on Wikidata or Orcid already have a unique identifier that can be used. | ||
How can I check if my import file is encoded in UTF-8?
- Open your file with the
Notepad app.
- Check the bottom right of the Notepad window.


- Save your file with the Save as option and change the encoding to UTF-8.
Data Aggregator functionalities
Can I upload images in the Data Aggregator?
There are two ways to have images associated to your records in DAGI.
- Insert the URL of your image (when deposited in a public repository) in the attribute associatedMedia –> see here below.
- Upload your pictures in ZIP files in the DAGI Media Store –> See the Guide section about Image Upload (available to DAGI users)
My images are already publicly available on a website, do I have to upload them too?
No, you don’t need the Media Store of DAGI if your images are already publicly available. What you can do is import the link to your images in the attribute associatedMedia.
associatedMedia : https://www.digitalis.uzh.ch/media/specimen/293/Z-000293332.jpg
I made a mistake when importing my data into the Data Aggregator, what do I do?
You can simply import a new file with the correct values and the same catalogNumber. The values in DAGI will be replaced. If you import empty values for an attribute in DAGI, then the attribute is emptied.
DAGI has a structure in three different layers (imported data, encoded data and validated data). For each of them, the history of all imported data is kept continuously. Therefore you can simply re-upload your correct dataset, do the correct mapping and encode it again. As long as your catalogNumber data is consistent, the rest is simply updated when importing a dataset with known catalogNumber values.
What are the different roles of the user profile on the Data Aggregator?
There are two different roles for user profile in DAGI:
- Collection Administrator
- can view, add, edit and remove users of their institution
- can view, add and edit datasets of their institution
- can publish datasets on GBIF.org
- Data Digitizer
- can view the datasets of their institution
- can import, encode, export and send records for Validation in the datasets of their institution
| Tasks | Collection Administrator | Data Digitizer |
|---|---|---|
| Users -view/add/edit/remove | ✅ | ❌ |
| Datasets -view | ✅ | ✅ |
| Datasets -add/edit | ✅ | ❌ |
| Records -import/encode/export/send for validation | ❌ | ✅ |
| Records -publish | ✅ | ❌ |
For all users and roles, the following rules apply:
🔸One institution can have more than one user / role
🔸One user is attributed to one and only one institution
🔸One user can have more than one role
🔸A user profile is specific to an individual and must not be shared with others
My institution does not have a Collection Digitizer yet, what can I do?
Please send an email to contact-swissnatcoll@infofauna.ch with your full name and institution name, so that we can add you as a Data Aggregator’s user.
How can I have more users in my institution?
Only Collection Administrators can add/edit/remove and assign roles to other users for their institution. To do this, go to the Administration page on DAGI (icon on the left side of your DAGI’s page) and click on 👤➕ Add user.
We consider that the Collection Administrator is fully responsible for the management of their institution page on DAGI. There is no need to ask for permission to add new users.
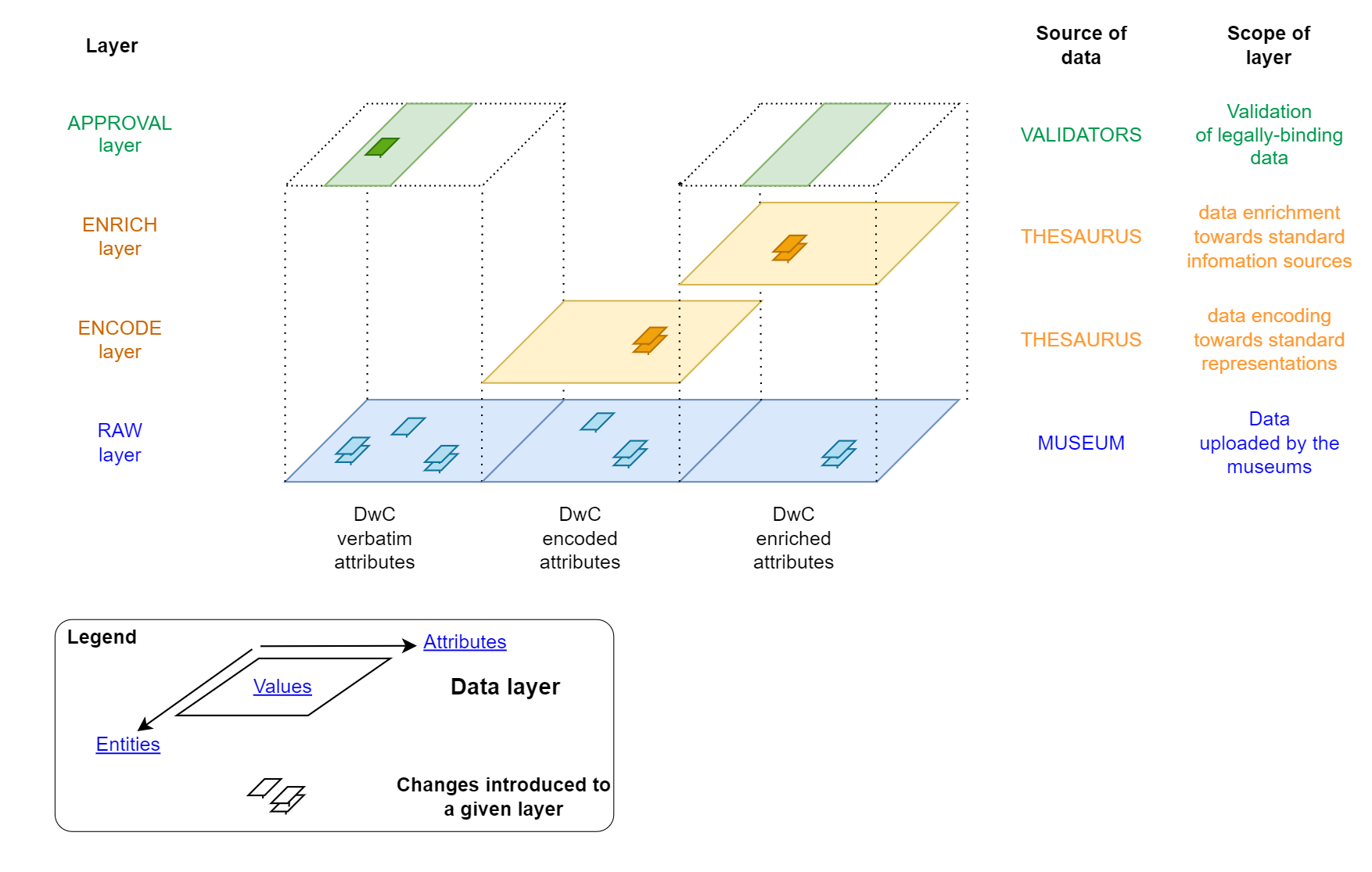
What are the different data layers of the Data Aggregator?
The data inside DAGI is organised in a table, with entities (records) in lines (= specimens with a unique catalogNumber value associated) and attributes in columns. The value is the information stored in a given attribute for a given record.
The values of a given entity can be added and updated in three different layers:
- The Raw layer: this layer contains the verbatim attributes and the interpreted attributes (encoded and enriched) imported in the import files. The import files come from the institutions, are uploaded by the institutions, are updated if a new import file contains entities (records) already present in the Raw layer.
- The Encoded layer: this layer is in two parts, the encoded part and the enriched part. For each encoded and enriched attribute, DAGI uses reference attributes (e.g. scientificName, locality, country, decimalLatitude/decimalLongitude, etc…) to fetch the corresponding values found in a set of thesaurii. The acquired values are then added to the corresponding entities’ attribute of that layer.
- The Validation layer: this layer concerns the validation of the data by the swiss Infospecies data centers, specifically in cases where sensitive data must be hidden or replaced by a less precise information (e.g. the coordinates of a rare and threatened species are replaced by their corresponding 10 km-square).
What does the Encoding do?
The Encoding process standardises important values of your data, and enriches your record with new standard information it did not have.
Here are the Thesaurii available in DAGI:
| Category | Resource | Field(s) used for query | Information encoded |
|---|---|---|---|
| GBIF Taxonomy | GBIF Species API | scientificName | taxonID, kingdom, phylum, order, class, family, genus, specificEpithet, scientificNameAuthorship, scientificName |
| Swiss Species | PICTIS | taxonID | taxonIdCH, acceptedNameUsage |
| Geo Reverse | OpenCage Geocoding API | decimalLatitude, decimalLongitude* | continent, country, countryCode, stateProvince, municipality, (if in CH: swissCoordinatesLv95_E/N and swissCoordinatesLv03_E/N) |
| Geo Forward | OpenCage Geocoding API | country, continent | continent, country, countryCode |
| GBIF IUCN Redlist | GBIF Species API | taxonID | iucn_redlist_category |
| Add Institution Code | GBIF Registry API (GRSciColl) | (Collection where encoding is done in DAGI) | institutionCode, institutionID |
| Relate Images | DAGI Media Store | Attribute selected during Image Upload | associatedMedia |
| Date Conversion** | DAGI internal code | a. eventDate b. day, month, year, endOfPeriodDay/Month/Year |
a. day, month, year, endOfPeriodDay/Month/Year b. eventDate |
*If decimalLatitude, decimalLongitude (=WGS84) is not informed, DAGI checks if swissCoordinatesLv95_E, swissCoordinatesLv95_N (=CH1903+/LV95) or swissCoordinatesLv03_E, swissCoordinatesLv03_N (=CH1903/LV03) are informed, and does the conversion to WGS84. The API is performed solely on decimalLatitude, decimalLongitude coordinates.
** eventDate value must be exactly ISO 8601-1:2019 standard YYYY-MM-DD
Examples of encoded data
| API Category | Source Data | Output Data = ENCODED | ||
|---|---|---|---|---|
| Attribute | Value (example) | Attribute | Value (example) | |
| GBIF Taxonomy | scientificName | Enydra anagallis Gardner | taxonID | 5402444 |
| kingdom | Plantae | |||
| phylum | Tracheophyta | |||
| class | Magnoliopsida | |||
| order | Asterales | |||
| family | Asteraceae | |||
| genus | Enydra | |||
| scientificName | Enydra anagallis Gardner | |||
| scientificNameAuthorship | Gardner | |||
| specificEpithet | anagallis | |||
| Swiss Species | taxonID | 5998041 | taxonIdCH | 22879 |
| acceptedNameUsage | Cerambyx miles Bonelli, 1823 | |||
| Geo Reverse | decimalLatitude decimalLongitude |
47.65545071 8.667665926 |
continent | Europe |
| country | Switzerland | |||
| countryCode | CH | |||
| stateProvince | Zurich | |||
| municipality | Benken (ZH) | |||
| swissCoordinatesLv95_E | 2692331.25671 | |||
| swissCoordinatesLv95_N | 1279034.48212 | |||
| swissCoordinatesLv03_E | 692331.25671 | |||
| swissCoordinatesLv03_N | 279034.48212 | |||
| Geo Forward | country continent |
Suisse Europe |
continent | Europe |
| country | Switzerland | |||
| countryCode | CH | |||
| GBIF IUCN Redlist | taxonID | 3188295 | iucn_redlist_category | EN |
Examples of data that can’t be correctly encoded
| DAGI Attribute | Value that can't be encoded | Cause | Recommendation |
|---|---|---|---|
| scientificName | "Rubiaceae Coffea liberica" | Presence of words corresponding to another rank than the species name (Genus + specific epithet + author) | Either import "Rubiaceae" or "Coffea liberica" |
| "example coming soon" | API call resulting in two options because two combinations exist with different authors | Add the author | |
| "Indet. ohne Angaben" | API call resulting in two options because two combinations exist with different authors | Either don't import these specimens with incomplete identification or choose a higher taxon rank (e.g. kingdom, phylum) | |
| eventDate | "01.01.2025" | Only the correct ISO 8601-1:2019 standard "YYYY-MM-DD" can be encoded by the Date Conversion | Separate your eventDate into day, month, year and use the encoding of DAGI or change the format to ISO 8601-1:2019 |
| "2025-1-1" | Only the correct ISO 8601-1:2019 standard "YYYY-MM-DD" can be encoded by the Date Conversion |
How do I most effectively open a csv file?
CSV files are quite tricky to open with Excel because the encoding of the file is forced by the system (usually ANSII), even if the file itself is UTF-8 encoded.
Here is the safest method to open any csv file:
- Open a new empty excel workbook.
- Click on Data in the Quick Access Toolbar (File - Home - Insert - Page Layout - Formulas - Data - Review …).
- Click on Get Data in the Ribbon (First element on the left of the Data ribbon).
- Choose From File and then From Text/CSV.
- A pop-up window opens, displaying the content of your CSV file, the corresponding encoding and the separation used. Make sure that the encoding format is “65001: Unicode (UTF-8)” for the File Origin.
- Click on Load at the bottom of the pop-up window
- Your data is now being loaded in your excel workbook and is now correctly displayed as a table.
How do I change the table format of my opened CSV file?
- Select your whole dataset (Crtl + A).
- Click on Table Design in the Quick Access Toolbar ( … Automate - Help - Table Design - Query).
- Click on Conver to range in the Ribbon (Second element, bottom line, on the left of the Table Design ribbon).
- Your data is now a usual excel sheet.
Special cases
My institution already has data on GBIF.org, how is this dealt with?
If some data in your database has already been sent to an Infospecies data center and/or GBIF Swiss Node and/or GBIF.org, please get in contact with GBIF Swiss Node before uploading this data in DAGI.
My institution has geological specimens (rocks and such), what have you planned about it?
Currently, DAGI and SwissNatColl hosted portal are mainly oriented for biological data (including fossils and paleontology). On the other side, GBIF.org does not support geological data.
The inclusion of the geological data of Switzerland is still in discussion, and no planning can be determined at the point where we stand at the moment.